
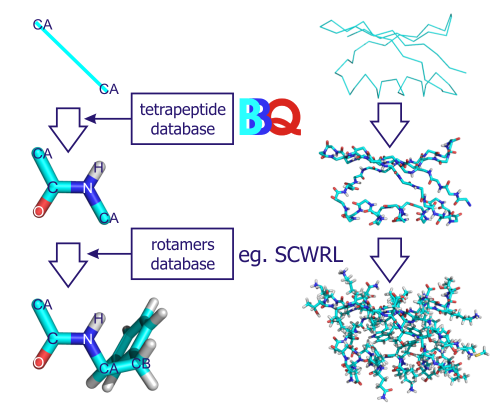
It is extremely important to understand the physical forces behind a peptide bond, as this allows scientists to design accurate, predictive models of three-dimensional protein structures. A -strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The backbone of a protein is composed of amide bonds, which are well solvated by (complexed with) the surrounding water. a bond angle of 180°) therefore, the peptide backbone must adopt specific conformations in order for atoms in peptide bonds to hydrogen bond with each other. A mistake in the translation process can lead to protein mis-folding, and in turn, disease. The backbone of a peptide chain is C C N where the middle C is the carbonly C O and C N is the peptide bond. Proteins can be as small as forty-four amino acids, or as large as thirty-five thousand. Peptide bonds are made within ribosomes during a process called «translation» to form polypeptides, which then undergo various molecular processing and modification, before folding into a three-dimensional shape, which we call a protein. For instance, there is currently much interest in antibody-drug conjugates these pair fragmented antibodies with pharmacologically active compounds in order to specifically target cancer tumors, among other things. The ability to predictably split peptide bonds is vital to a number of different fields of study. Reversing a peptide bond without an enzyme is extremely difficult, thus this process is usually mediated by an enzyme called a protease, such as subtilisin, which is frequently added to laundry detergent to cleave many protein contaminants.
It may therefore be counterintuitive to learn that peptide bonds are quite stable kinetically: the lifetime of a peptide bond in aqueous solution is approximately 1000 years.
#Peptide backbone of a protein free#
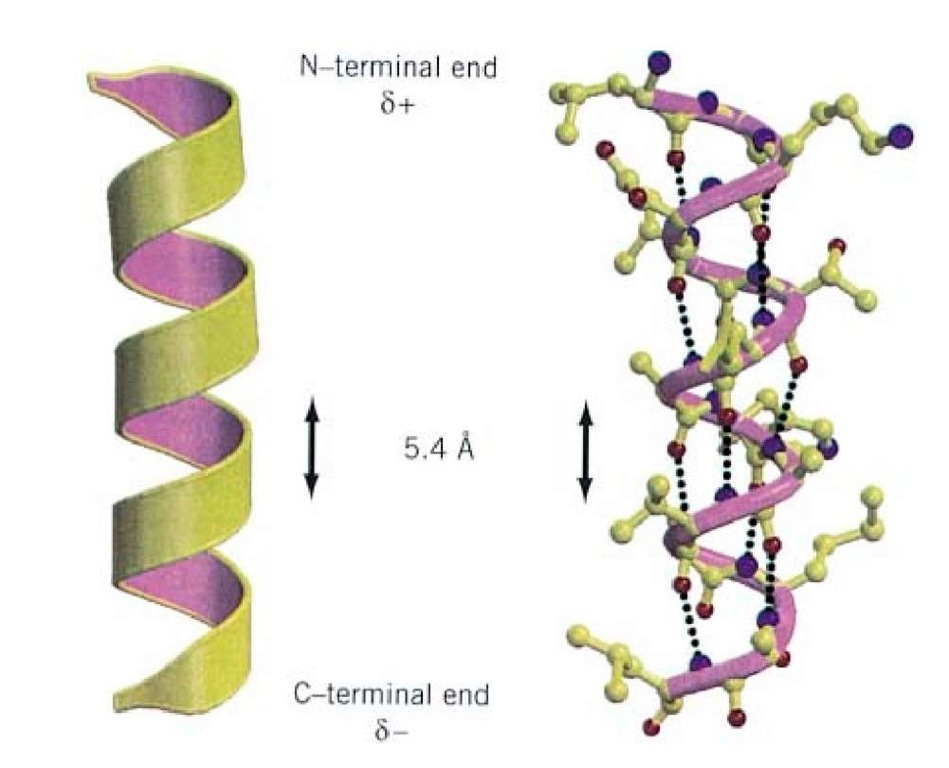
Hence, the biosynthesis of a peptide bond requires an input of free energy. One interesting thing to note is that the equilibrium of this reaction lies on the side of hydrolysis rather than synthesis. First, surfacecomplementing protein fragments, which we dub interface seeds, are generated around the target protein using TERMs. The backbone design process proceeds in three steps. For most peptides the cis-form is about 1000 times less stable than the trans-form. We devised a method to generate surfacecomplementing peptide backbone structures from TERMs. In such cases, the cis form is more stable than usual since the proline side-chain offers less of a hindrance. The alpha helix is stabilized by hydrogen bonding interactions between peptide backbone carbonyl (CO) oxygen atoms and peptide backbone amide groups 4. However, cis forms can occur in peptide bonds that precede a proline residue. In naturally occurring peptides most peptide bonds are in the trans configuration.


 0 kommentar(er)
0 kommentar(er)
